See the people, processes, and equipment fueling Li’s internationally recognized research program
By Jill Sakai
The Li Research Group has discovered more than 300 novel neuropeptides, been featured on the cover of more than 20 scientific journals, and produced hundreds of publications helping the scientific community to build their understanding of lipids, peptides, proteins, and other molecules in the context of cancer, neurodegenerative disease, and more.
This highly productive lab is run by Professor Lingjun Li, Vilas Distinguished Achievement Professor and Charles Melbourne Johnson Distinguished Chair in Pharmaceutical Sciences at the University of Wisconsin–Madison School of Pharmacy. Li, who is also a professor with the UW Department of Chemistry and mentors Chemistry, Biophysics, and Pharmaceutical Sciences graduate students, has been named one of the world’s most influential analytical scientists three times.
With a strong background in analytical chemistry and neuroscience, she built a research program centered around discovering the surprisingly extensive array of small signaling molecules called neuropeptides in the nervous system, and analyzing their roles in behavior and health.
“Our lab uses bioanalytical chemistry to address challenges in basic and biomedical research,” Li says.
So what does this field-leading work look like? Let’s take a look at the Li Research Group in action.

The Li Research Group’s focus is on creating tools and technologies using mass spectrometry, a technique that distinguishes molecules based on their weight, shape, charge, and other physical properties. This powerful timsTOF fleX MALDI-2 (Matrix Assisted Laser Desorption/Ionization) instrument is one of several different mass spectrometers available in the School of Pharmacy, which together enable a wide variety of types of studies. One advantage of this technique is that it gives a readout of all the molecules present in a sample, making it a powerful tool for discovering unexpected or novel chemical signatures.
Here, postdoctoral fellow Hua Zhang is loading the MALDI-2 instrument with a sample plate containing a thin slice of brain tissue from a mouse model of Alzheimer’s disease. The instrument is capable of scanning the tissue section to map the spatial distribution patterns of the biomolecules in the brain tissue based on their molecular weights and characteristics. Zhang is studying how the distribution of phospholipids changes in neurodegenerative disease.
Li describes imaging mass spectrometry, such as Zhang’s Alzheimer’s disease project, as “the new frontier” of mass spectrometry capabilities. The lab was recently awarded a new five-year R01 grant from the National Institutes of Health to study patterns of biomolecules localized to specific brain regions.
“A lot of our effort is trying to improve the sensitivity, the spatial resolution, and the type of molecules that we can see,” Li says.
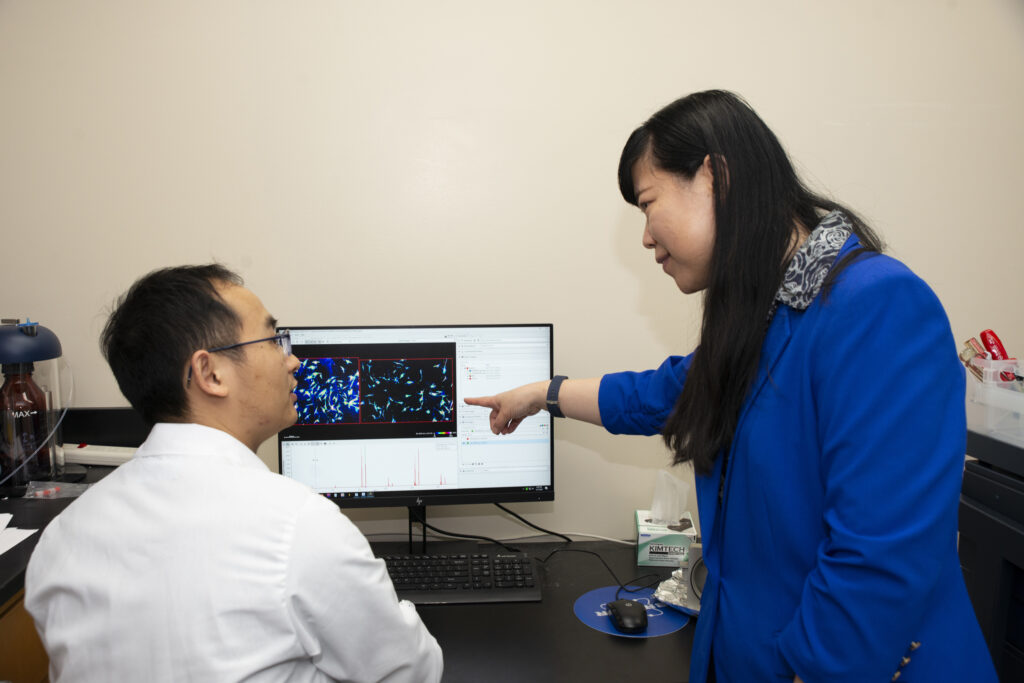
The lab also works on several types of cancer. Here, Li (right) talks with Zhang about phospholipids produced in pancreatic stromal cells. These images, also generated from the MALDI-2 instrument, are part of a project to better define chemical signatures in the tissue surrounding a tumor and how those signatures might relate to the tumor’s ability to metastasize. The researchers can study how the phospholipids change under different conditions and in response to drug treatments.
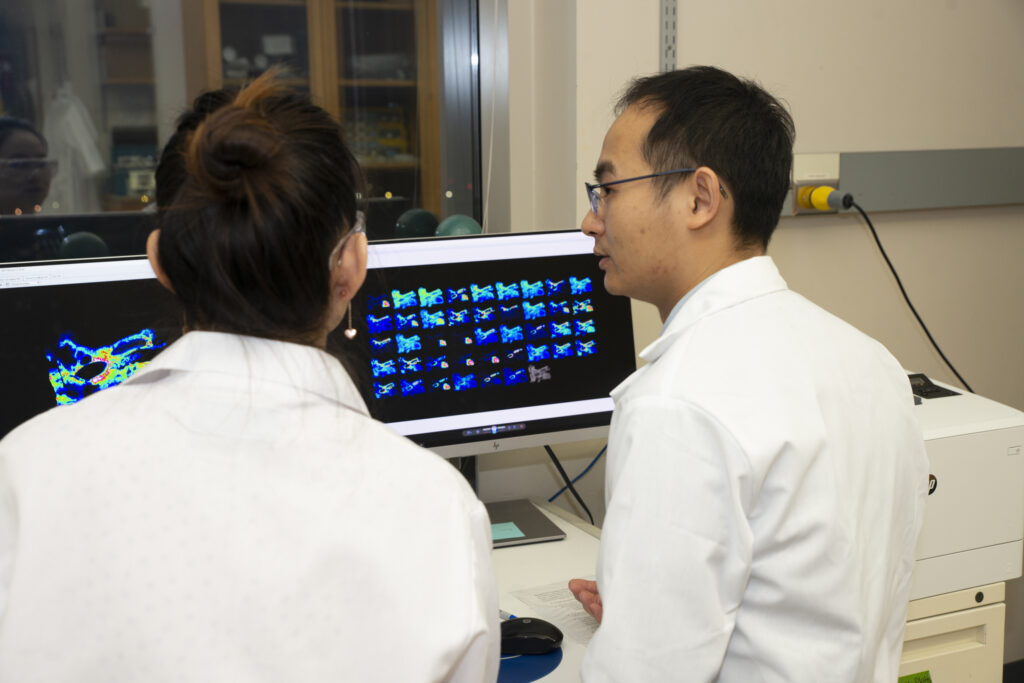
Zhang (right) and postdoctoral fellow Min Ma analyze mass spectrometry images of a human laryngeal cancer biopsy, collected as part of a collaboration with Nathan Welham in the Department of Surgery. Zhang has developed new methods that can improve the detection and separation of a type of carbohydrate called N-glycans. These molecules have been difficult to detect in cancer tissue samples, but they may yield useful biomolecular signatures for studying disease.
Many disease processes involve altered levels or regulation of a host of molecules — peptides, glycans, proteins and more — individually or in networks, Li says.
“We develop mass spectrometry-based technologies to try to identify and quantify those differences, even at very small scale,” she says. “The more complete picture you can get of a biological system, the more insight you can gain into understanding complex diseases.”

One technique the laboratory has advanced is ion mobility mass spectrometry. This method adds an extra separation step that helps distinguish among very similar molecules such as isomers, which contain the same atoms and are the same weight but have slightly different shapes or chemical configurations.
“Some of these subtle differences may turn out to be important biomarkers for disease,” Li says. For example, most of the amino acid building blocks our bodies use to assemble proteins adopt an “L” configuration, but Li’s team has found elevated levels of the mirror-image “D” amino acids in the brain tissue of a mouse model of Alzheimer’s.
Andrew Zhu, an analytical chemistry graduate student in the Li Research Group, is developing methods to be able to study peptide and fatty acid isomers. Here, he is processing high-resolution ion mobility data to look at isomers of different fatty acids, which are essential molecular regulators of many biological processes.
The improved separation that comes with ion mobility mass spectrometry also generates huge quantities of raw information. Those datasets require a lot of highly skilled processing for the researchers to identify molecules of interest.
“A lot of our work involves trying to make sense of those data,” Li explains.
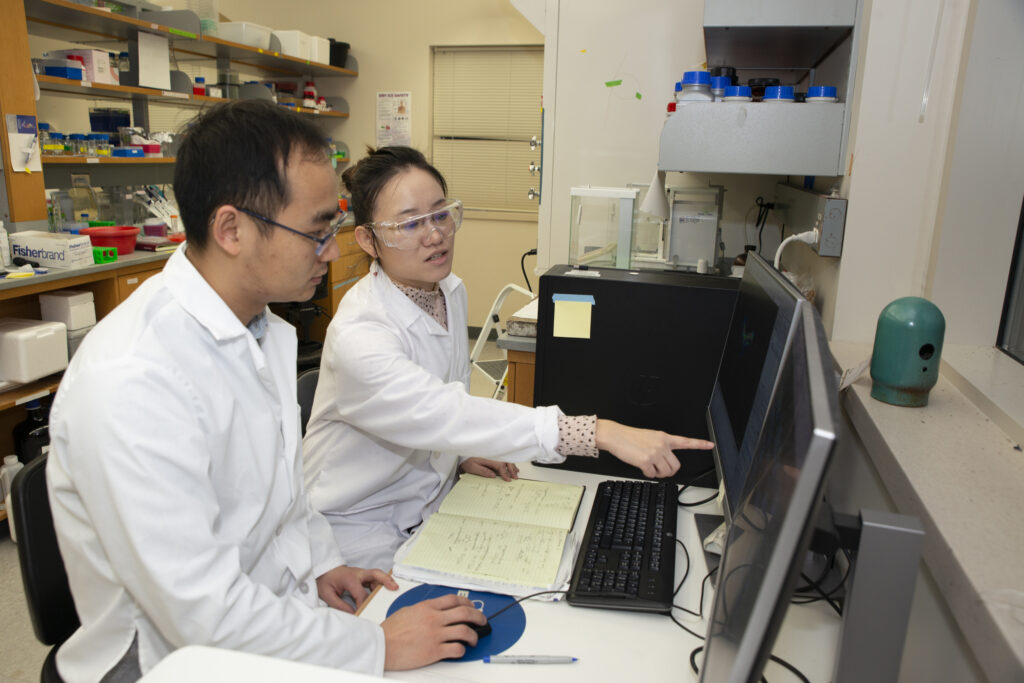
Zhu (right) and postdoctoral fellow Shuling Xu are analyzing data to quantify levels of D- vs L-amino acids in a sample. While in the lab, many students also learn to code to be able to write programs that streamline data processing.

Another large component of the team’s work is preparing the samples for the mass spectrometry analyses. The specific steps — and how long they take — depend on the type of samples and can take days or even weeks, Li explains.
Some projects require pre-treatment to digest tissues or break large proteins into smaller bits. Others require synthesis of chemical tags to label a subset of molecules for easier identification or more accurate quantification in the end dataset. Often, an enrichment step is necessary to boost molecules present at very low levels and make detection more reliable. Here, Pharmaceutical Sciences graduate student Feixuan Wu uses a micropipette to process a sample for mass spectrometry analysis.

Wu (left) and analytical chemistry student Dylan Tabang talk through needed sample preparation steps. There's a lot of peer training and collaboration among students and postdoctoral fellows in the lab, Li says. These two students are working with a microcentrifuge to prepare samples for a project studying a type of protein modification called O-glycosylation and comparing it between healthy and diseased pancreatic tissues.

Ting-Jia Gu, a Pharmaceutical Sciences graduate student, records lab notes about a recent project. Gu develops and synthesizes chemical tags to use in quantifying amounts of various phospholipids and proteins. Planning, executing, and documenting all the steps involved in chemical synthesis requires meticulous record-keeping.

Peng-Kai Liu, a biophysics graduate student, works with a piece of specialized equipment called a rotary evaporator, which is used in chemical synthesis. Li calls Liu, Gu, and Pharmaceutical Sciences graduate student Zicong Wang (not pictured) “our synthetic chemistry team,” responsible for driving much of the lab’s work making chemical tags to label various biomolecules. Student interests often end up sparking new research directions and collaborations, Li notes.
“Over the years, our research program has grown quite a bit because the tools that we develop allow us to look at more types of biomolecules in a broader range of applications,” she says.

A close-up of the rotary evaporator shows the glass chambers used to heat and concentrate samples. After running a chemical synthesis reaction, Liu uses this “rotovap” to purify the desired compound so he can use it to make a chemical tag.
“This is actually a pretty tedious process,” Li says.

The Li Research Group pursues a mix of projects exploring fundamental neuroscience questions in addition to their translational and clinical projects on Alzheimer's disease and pancreatic cancer. Chemistry graduate student Wenxin Wu and other lab members use crustaceans, including the lobsters shown here, as model systems to study how small signaling proteins called neuropeptides change during feeding or other behaviors, or in response to environmental changes such as temperature or oxygen levels. Wu also helps care for the animals, which are housed in a specially designed aquatic facility in the building.
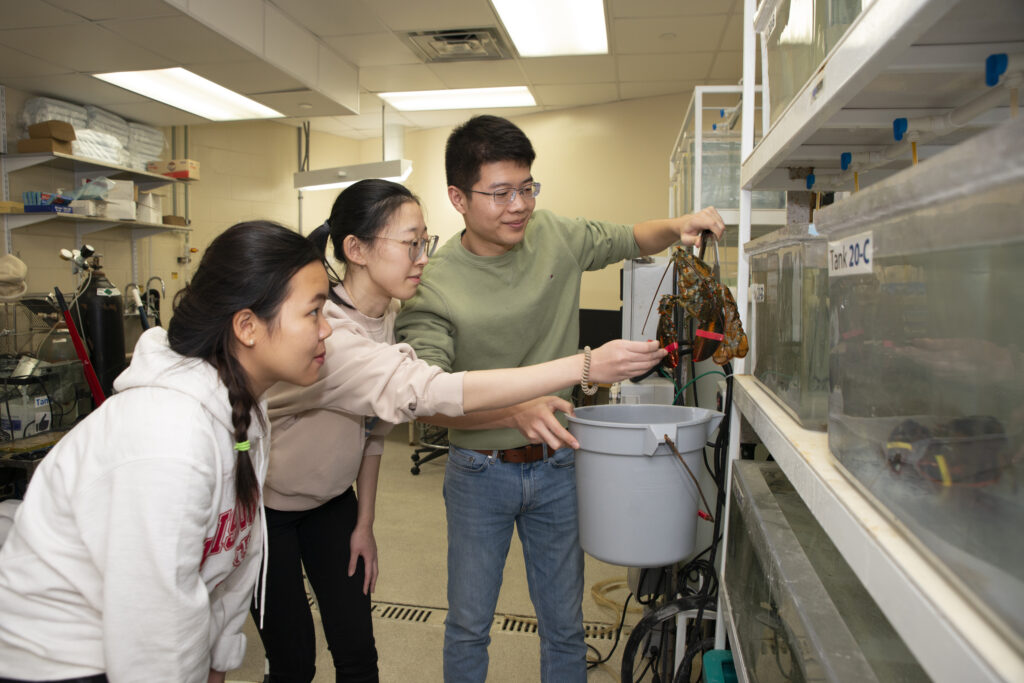
Wu (center) and Pharmaceutical Sciences graduate student Gaoyuan Lu (right) are training new graduate student Huong Tran to work with the lobsters.
Early in her career, Li became fascinated with mass spectrometry’s capabilities and its potential as a tool for biological discovery. Crustaceans offered the advantage of a relatively small, well-defined nervous system.
“When I started, there were about 20 known neuropeptides,” Li says. “But when we applied mass spectrometry, just to look at this small neural circuit, we found hundreds of unknown peaks.” The discovery of that unexpected complexity “revolutionized people's thinking about neuropeptides” and the multitude of roles they may play in the body.
Mass spectrometry is still uncovering surprises in Li’s lab. Lu has now found several thousand peptides and neuropeptides in lobsters, including some that contain the unusual “D” type of amino acids. It’s exactly the type of discovery that’s likely to spur a new phase of Li’s research program.
“A lot of our tool development is propelled by our curiosity, the need to understand the biology more in depth,” says Li. “The biological problem drives method development, then the new technology development enables more, broader biology applications in an ever-expanding cycle. That’s the evolution of our research.”

The Li Research Group
Front Row (left to right): Jericha Mill, Zexin Zhu, Hua Zhang, Dylan Tabang, Min Ma, Lingjun Li, Wenxin Wu, Hannah Miles, Devin Tomlin, Feixuan Wu
Middle Row (left to right): Graham Delafield, Cameron Kaminsky, Angel Ibarra, Ting-Jia Gu, Danqing Wang, Haiyan Lu, Shuling Xu, Ashley Phetsanthad, Kelly Lu, Huong Tran, Lauren Fields, Ching-Yuan Yang
Back Row (left to right): Malik Ebbini, Elliot Patrenets, Haoran Zhang, Gaoyuan Lu, Zhijun Zhu, Eric Chiang, Peng-Kai Liu, Bin Wang, Zicong Wang, Yuan Liu
Not pictured: Olga Riusech, Peng-Hsuan Huang, Jingwei Zhang, Tina Dang, Thao Duong, Mitch Gray