9
March

Alum Ian Miller designs ‘smart toilet’ to capture health information from drug metabolism to exercise
By Andrea Mongler
When Ian Miller (PhD ’18), was a graduate student at the University of Wisconsin–Madison School of Pharmacy, he kept an open mind about his future career. Still, the possibility that he might devote years to designing a toilet never occurred to him. But as it turns out, his experience at the School of Pharmacy put him on a path to do just that.
Miller and his colleagues aren’t designing just any old toilet. Instead, they’re working on a “smart toilet” they hope can be used to monitor and improve health. Specifically, the toilet would collect and analyze urine, which contains metabolites that provide clues about a person’s daily life and their health.
“We want to use the abundance of metabolites in urine to learn about health and lifestyle in a non-invasive and passive way,” says Miller, who is now a data scientist in the Coon Research Group at UW–Madison, where he’s affiliated with the Department of Biomolecular Chemistry at the School of Medicine and Public Health. “Similar to the way that a smartwatch collects heart rate data, we’re envisioning using urine to understand daily metabolic health.”
Ultimately, Miller and his colleagues hope their work will help advance the field of personalized medicine. It’s a big goal, but they started small — by collecting and analyzing their own urine.
“If our technology was broadly available for passive data collection, we could provide pharmacists feedback in real time and remotely on how and how quickly patients are metabolizing drugs so that a dose could be updated or optimized or a drug regimen could be changed altogether.” –Ian Miller
Uncovering your metabolic “fingerprint”
As part of a small pilot study last year, Miller and Josh Coon, professor of chemistry and biomolecular chemistry at UW–Madison, collected their own urine for a 10-day period. In total, they collected 110 samples and analyzed them using technology called gas chromatography and mass spectrometry.
Gas chromatography allows researchers to separate complex mixtures so that the compounds they contain can be presented individually to a mass spectrometer. The mass spectrometer, in turn, identifies the chemical structures that make up these compounds.
In other words, it shows researchers what’s inside the samples. In Miller and Coon’s case, their urine samples revealed a lot about their daily lives. They were able to identify biomarkers connected to their coffee and alcohol consumption, for example, and they could tell when they’d taken acetaminophen (e.g., TYLENOL) — which is especially relevant to pharmaceutical science and personalized medicine.
“With the ability to detect drugs and drug metabolites in urine, you could tailor dosages based on how quickly someone is metabolizing a drug, which is sometimes difficult to predict from genome sequence data alone,” Miller says.
Miller and Coon also used wearable technology and smartphone apps to track their heart rates, number of steps, calorie consumption, and sleep patterns. They integrated this data with the metabolomic data from the urine samples, which helps provide a more complete snapshot of someone’s health. For example, it may be possible to better understand how caffeine consumption affects sleep by measuring caffeine metabolites in urine and integrating that data with sleep data collected by a wearable device or mobile app.
In addition, their analysis showed clear differences between each participant’s urine samples. In a paper published in the Nature journal npj Digital Medicine in November 2019, the researchers noted that each participant had “a distinct baseline metabolic fingerprint.”
These “fingerprints” have implications for disease prevention. Metabolites linked to many diseases and conditions can be identified in urine, potentially providing an opportunity to prevent them. Knowing someone’s baseline metabolic fingerprint makes this possible on a highly individualized level.
“Most routine clinical measurements are made on an infrequent basis — maybe annually as part of a regular checkup — and check only a few things, such as blood lipid levels and fasting glucose levels, which are then compared against the entire population,” Miller says. “It would be more meaningful to compare deviations from an individual’s baseline instead of a discrete difference from the population average in a measurement taken infrequently.”
“With the smart toilet, we could theoretically monitor and diagnose a diverse array of diseases, but we will probably focus our efforts on diseases affecting the kidney, liver, and immune function.” –Ian Miller
Processing samples using the toilet
The next step for Miller and his colleagues is to collect data from more participants — and that’s where the smart toilet comes in. As Miller can attest, collecting your own urine repeatedly for several days is a bit of a burden, so expecting a large group of participants to do so reliably isn’t particularly realistic.
That’s why the research team is designing a toilet, or likely a device that can be installed in a traditional toilet, that can collect and analyze urine samples, which will take the burden off of study participants.
“With the smart toilet we’re designing, people won’t have to change their behavior at all,” Miller says. “They’ll simply use the toilet as usual, and we’ll have access to continuous metabolic health data.”
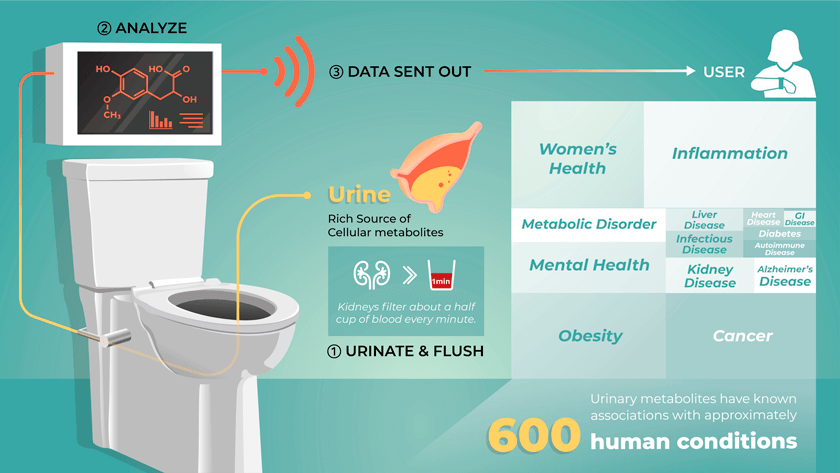
The plan is to connect the toilet directly to a portable mass spectrometer. A urine collection system in the toilet would send urine to the mass spectrometer, which would then analyze the samples before they’re flushed as usual. Data from the analysis will likely be accessible through a smartphone application.
The research team plans to install the toilet in their research building on campus and recruit dozens of participants for this larger study to test the toilet. Miller says he and his colleagues are aiming to have a product on the market within five years.
“With the smart toilet, we could theoretically monitor and diagnose a diverse array of diseases, but we will probably focus our efforts on diseases affecting the kidney, liver, and immune function,” he says. “From a clinical standpoint, such a device may be most immediately relevant to patients with diabetes since they are at increased risk of kidney and liver disease.”
“Dr. Kwan has profoundly influenced the way I think about scientific problems and the diverse data surrounding them.” –Ian Miller
The pharmaceutical link
In addition to helping plan the pilot study, Miller interpreted the mass spectrometry data and integrated it with data from the wearable devices and smartphone apps. His experience at the School of Pharmacy prepared him for this work in more ways than one.
He first worked with mass spectrometers as a project assistant in the School of Pharmacy’s Analytical Instrumentation Center. And during his PhD project in the lab of Jason Kwan, assistant professor in the School’s Pharmaceutical Sciences Division, Miller gained experience with bioinformatics, data science, and programming.
“The informatics and software skills I acquired in the Kwan Lab have been incredibly valuable to me in interpreting metabolomics data for the smart toilet project,” Miller says.
He says he was the first graduate student to work in the Kwan Lab, and Kwan’s impact on him persists.
“Dr. Kwan has profoundly influenced the way I think about scientific problems and the diverse data surrounding them,” Miller says. “I’m indebted to him for the skills and insights that will forever shape my perspective and professional trajectory.”
Miller’s pharmaceutical background and training complement the skill sets of his co-investigators on the smart toilet project. The interdisciplinary team also consists of chemists, biologists, physicists, engineers, and bioinformaticians. In fact, Miller says, the work requires an interdisciplinary team.
Miller and his colleagues hope that once they finish building — and then testing — the smart toilet, they can adapt the technology for use on a much larger scale. Miller is excited about the potential implications of this, including for the field of pharmaceutical sciences.
“If our technology was broadly available for passive data collection, we could provide pharmacists feedback in real time and remotely on how and how quickly patients are metabolizing drugs so that a dose could be updated or optimized or a drug regimen could be changed altogether,” he says.
Just how broadly available the technology may eventually be is anyone’s guess. Miller says he envisions the smart toilet becoming a common feature in home bathrooms, allowing people to easily monitor their health from the comfort of their own homes.
“We are still working on market analysis, so our go-to-market strategy will continue to be refined based on feedback from potential users and early adopters,” he says. “But we imagine that, initially, this technology may be of particular interest to the people using wearable devices and smartphone applications to get regular digital readouts of their health.”
Miller also noted that the technology could be implemented in nursing homes, hospitals, and athletic facilities.
“I like the idea of enabling health monitoring through a toilet,” he says. “If we are successful in implementing such a device at scale, I believe we could really improve modern health care.”
Learn more about the Pharmaceutical Sciences PhD Program at the School of Pharmacy.