10
December

Armed with two new grants, School of Pharmacy Assistant Professor Jason Peters continues his quest to find new targets for the hardest-to-treat bacterial infections
By Nicole Sweeney Etter
It’s been called “the slow-moving pandemic.” Bacterial infections are one of the leading causes of death globally, and as more pathogens become resistant to antibiotics, the threat to human health is expected to grow.
But there are reasons for optimism as researchers continue to chip away at the problem, says Jason Peters, an assistant professor in the University of Wisconsin–Madison School of Pharmacy’s Pharmaceutical Sciences Division.
“I think that we have the research tools in place to make a lot of progress here,” says Peters, who recently won two new grants to accelerate his work on antibiotic resistance.
“We’re trying to understand the interactions between bacterial genes and antibiotics for the sake of understanding how antibiotics work, but also designing new, better antibiotics.”
—Jason Peters
The first is a $1.9 million grant from the National Institute of General Medical Sciences to study bacterial gene networks, which could reveal new targets for antimicrobial compounds. For the second grant, Peters’ lab is one of 25 teams working on the multi-institutional, $104 million grant, funded by Advanced Research Projects Agency for Health (ARPA-H), that aims to develop a new diagnostic tool for bacterial infections and identify new targets to help defeat the growing problem of antibiotic resistance.
Together, the projects span the spectrum from foundational basic science to translational research that will more quickly reach patients. But both approaches are important in the fight against antibiotic resistance.
Understanding bacterial gene networks
The goal of Peters’ first grant is to understand how genes are functioning in bacteria. Historically, researchers have studied bacterial genes as isolated units, examining the 4,000 or more genes in a typical bacterial genome one at a time.
“That’s a great way of looking at things, but it’s really slow,” Peters notes. And in the race against antibiotic resistance, time is essential.
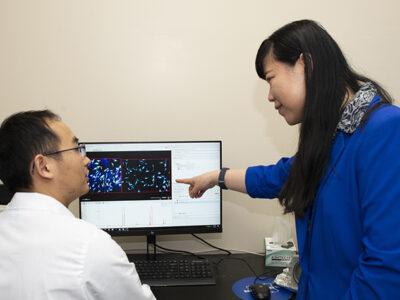
By using CRISPR genetic tools, Peters can perturb all the genes simultaneously and then measure the results.
“We’re trying to understand the interactions between bacterial genes and antibiotics for the sake of understanding how antibiotics work, but also designing new, better antibiotics,” he says. “And we’re also trying to understand how genes work together.”
That approach could reveal new vulnerabilities in bacteria.
“In some ways, genes can actually hide their functions,” he explains. “They get together with their friends that have the same activities, and they do the thing that they’re doing — in this case, allowing the cells to become antibiotic resistant. And now you can’t tell that those genes were important for antibiotic resistance because when you perturb one of them at a time, their friend just takes over that role. So we’re perturbing multiple genes at once to look for how the genes are connected and how they accomplish similar functions.”
Studying 4,000 genes in pairs would mean analyzing 16 million possible combinations, and the number gets even bigger if researchers want to examine larger combinations of genes. While it sounds mind-boggling, CRISPR technology makes it possible.
“It’s really a combination of factors and the scale that is pushing this research forward. That’s where the major innovation is,” Peters says. “And you can learn things at that scale that you can’t learn from individual experiments.”
Peters will also measure the response of cells when exposed to different antibiotics.
”We’re going in with an expectation of delivering something translational — not just a promise of having information that could one day be used translationally.”
—Jason Peters
“The more antibiotics we look at, the more likely we are to find genes that are important for one antibiotic but not for another, or genes that are generally important for surviving antibiotics,” he explains. “By not narrowing down our scope ahead of time, it allows us to see a complete picture.”
Over the five-year grant, the Peters Lab will learn more about the fundamental mechanisms of how bacterial cells function — insights that could inform more applied approaches for countering antibiotic resistance, including the ARPA-H project.
“A lot of the translational work that will be done later is powered by some of the observations that we’ve made at a very basic level,” he says. “You have to build this infrastructure in order to make those discoveries.”
Collaborating to improve patient care
The multi-institutional project funded by ARPA-H is led by Johan Paulsson, a professor of systems biology at Harvard Medical School. Paulsson sought out Peters as a collaborator because of his expertise in developing and implementing CRISPR tools to understand bacterial genetics and antibiotic functioning.
The ambitious effort aims to uncover the mechanisms driving antimicrobial resistance and to create a new diagnostic tool to identify pathogens at the point of care in hospitals.
“It’s pretty surprising how difficult it can be to detect specific pathogens that are causing infections,” Peters explains. “Usually it’s a drawn-out process that involves culturing bacteria and kind of a test-and-hope-it-works strategy with antibiotics. One of the approaches that this grant is trying to push forward is ways of uniquely identifying certain pathogens and getting that information out more quickly.”
The grant will also explore how bacterial genes interact with established antibiotics and new potential antimicrobials.

Researchers typically study how antibiotics kill bacteria by simply measuring the presence of bacteria after treatment. In this study, researchers will be able to explore the effects of antibiotics on the cellular level to see how individual cells change shape, indicating how sensitive those cells are to a particular antibiotic. That could provide insights into how subpopulations of cells develop antibiotic resistance, which can eventually lead to antibiotic failure.
“We’re basically taking the incredible scale of our genetic methods and combining it with the incredible scale of Johan Paulsson’s microscopy techniques. And now we’re going to be able to look at the behavior of bacteria at an unprecedented scale and learn a lot about how antibiotics work at the single cell level,” he says. “And that’s really important because a lot of antibiotic resistance phenotypes occur at the single cell level.”
By the end of the five-year grant, researchers hope to have identified new antimicrobial compounds and have a prototype of a new diagnostic device ready for testing in the hospital environment. The overall pace of the project is exceptionally fast, Peters notes, with the goal of creating something ready to be used for patient care.
“It’s certainly nothing like I have worked on before,” he says. “It’s really an all-hands-on-deck kind of effort, with all kinds of different expertise. And we’re going in with an expectation of delivering something translational — not just a promise of having information that could one day be used translationally.”