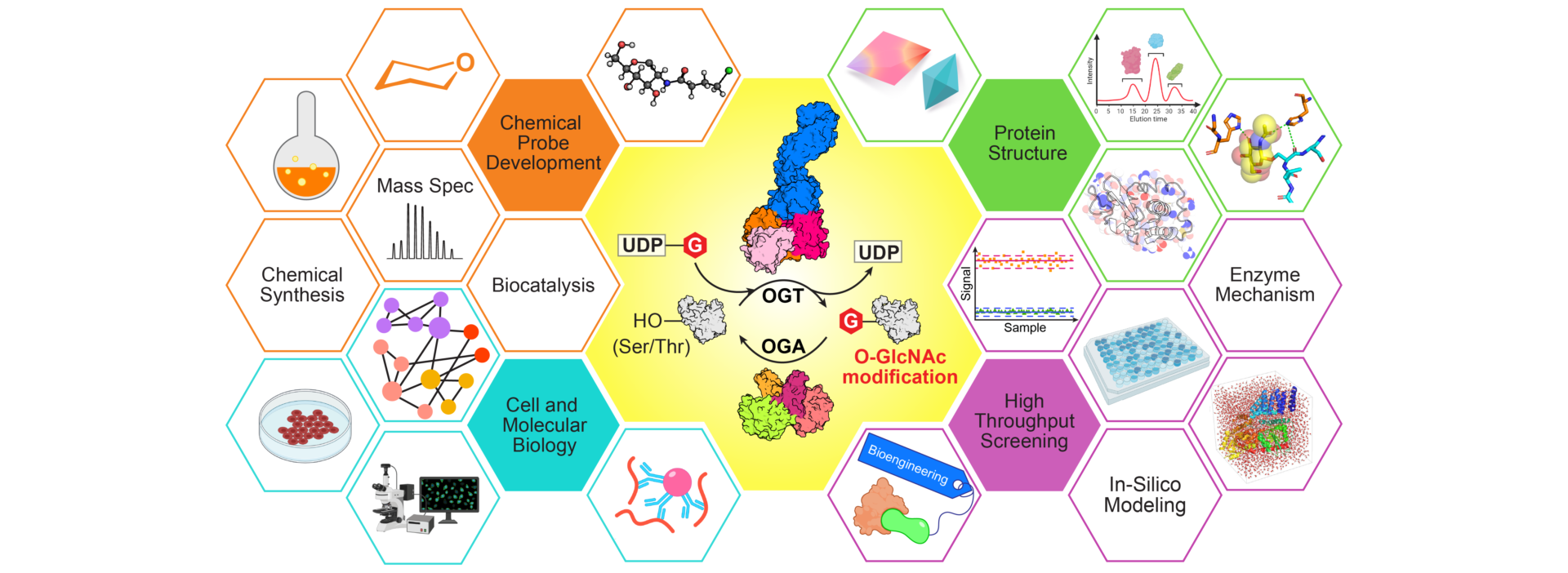
The overarching goal of our research program is to understand the molecular mechanisms and biological functions of protein glycosylation, especially O-GlcNAcylation. This unique modification dynamically regulates a wide variety of physiological and pathological processes. However, the mechanism underlying its dynamic regulation and aberrant function remains largely unknown. The Jiang Research Group has been active in developing innovative tools, determining novel protein structures, and uncovering regulatory pathways to characterize the functions of O-GlcNAc cycling enzymes (OGT and OGA), and design new strategies to control the level of this modification for drug discovery.
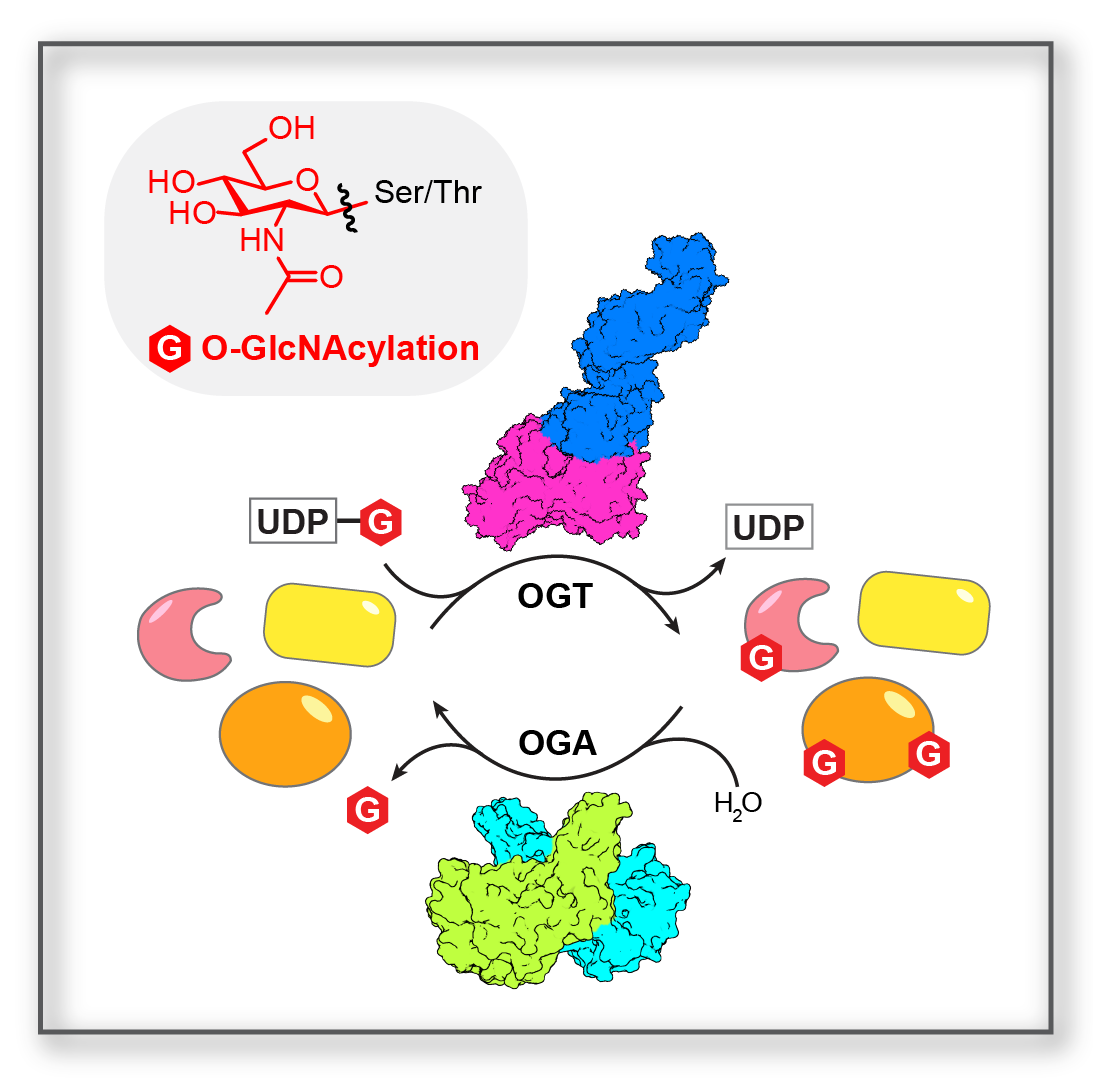 Protein Structure and Mechanism Protein Structure and Mechanism |
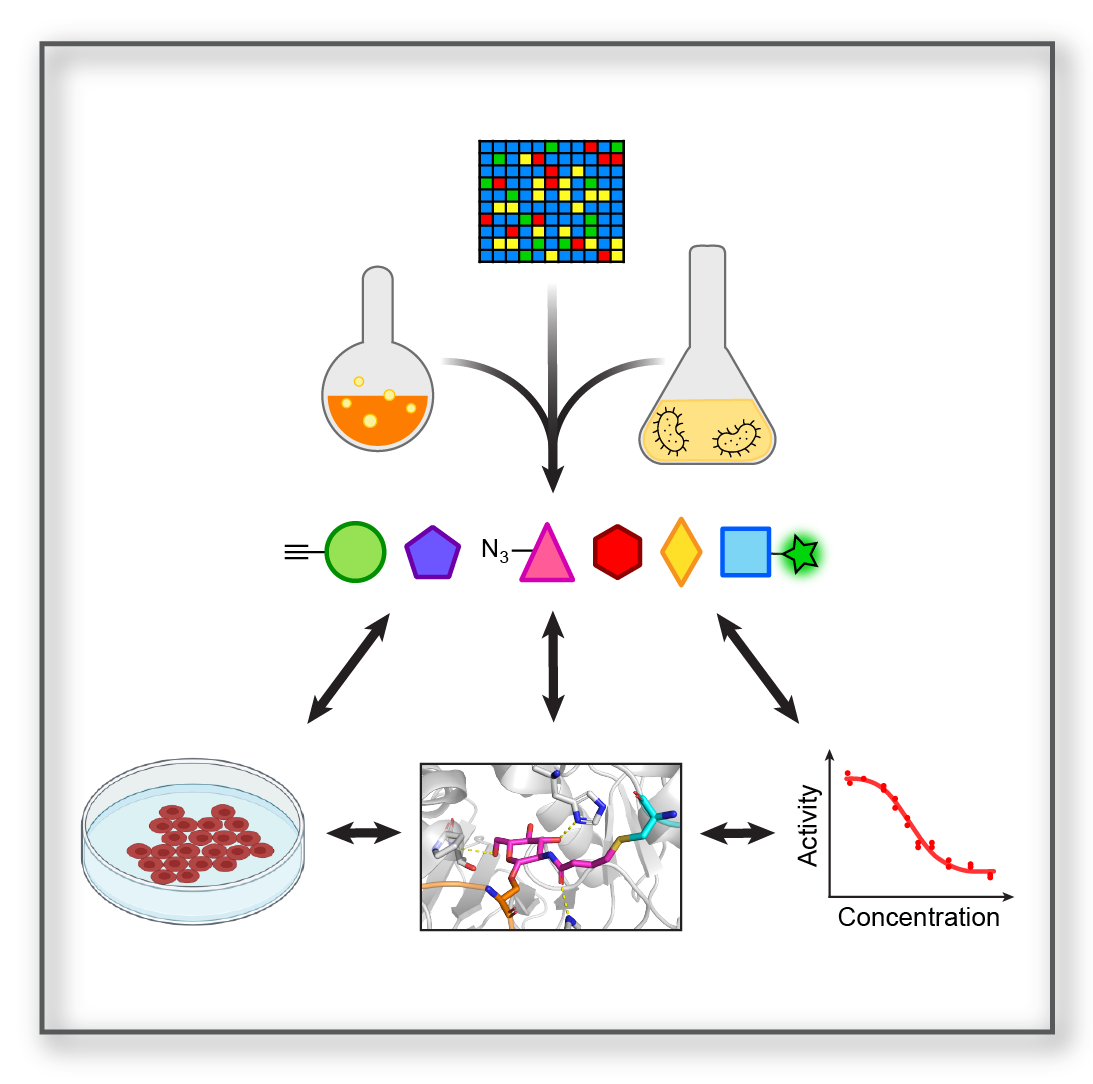 Chemical Probe Development Chemical Probe Development |
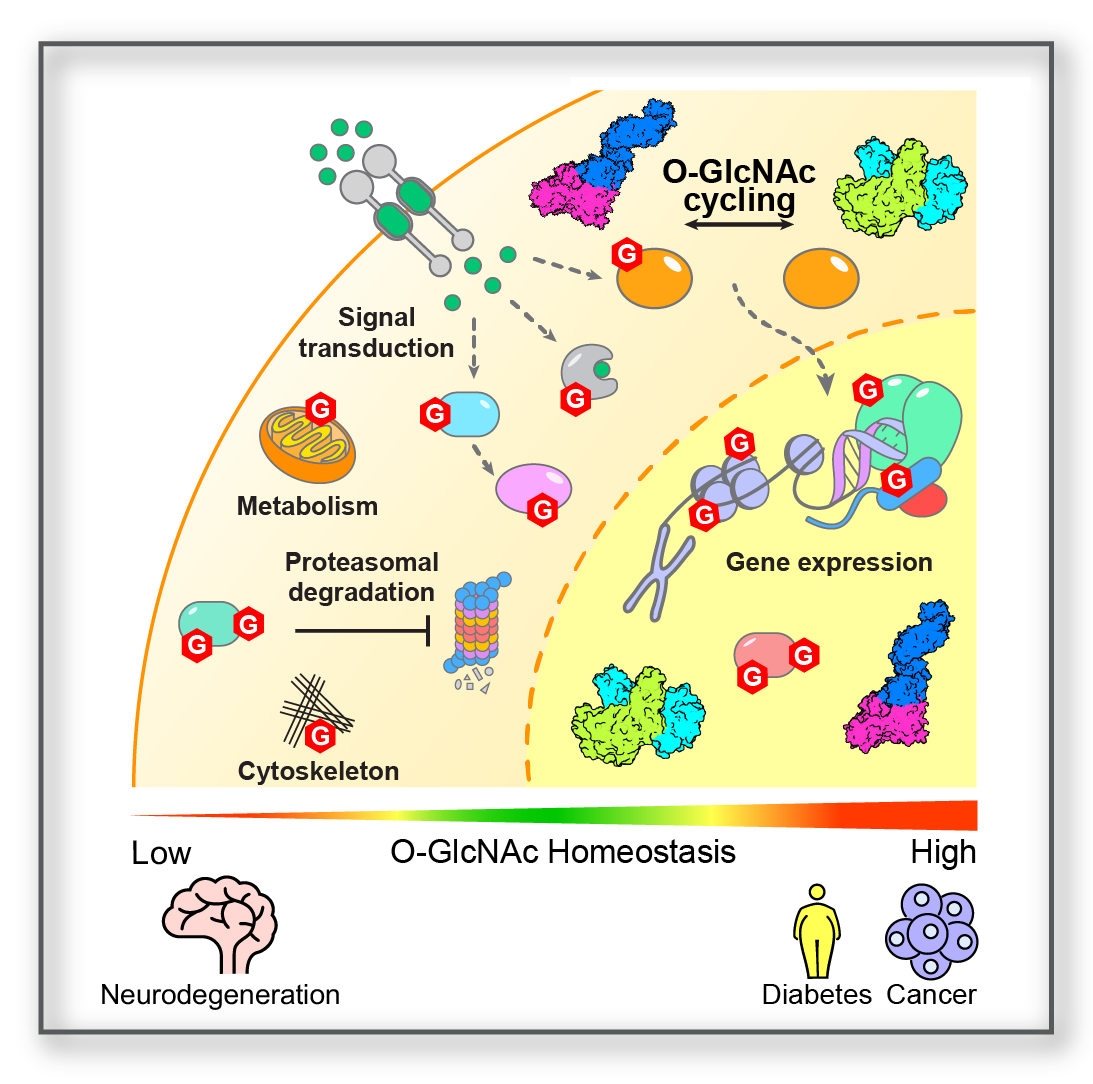 Regulation of Cellular Functions Regulation of Cellular Functions |