13
September

Postdoctoral scientist Sarah Neuman and Professor Arash Bashirullah find bridge-shaped proteins that channel the flow of essential lipids inside cells
By Jill Sakai
An organism’s genes contain all the information needed for it to grow and function. When something goes wrong during development or disease, that often signals a defect in one or more genes.
Although the human genome was fully sequenced 20 years ago, scientists still don’t know the function of the vast majority of human genes. Studying previously uncharacterized genes in simpler organisms can help identify their function. But as developmental geneticist Sarah Neuman has learned, it often takes extensive sleuthing to connect visible defects or disease states to the underlying function of the associated gene.

As a graduate student, Neuman worked with Arash Bashirullah, professor in the University of Wisconsin–Madison School of Pharmacy’s Pharmaceutical Sciences Division, to understand what was going wrong in a set of genetically mutated fruit flies that were abnormally small and died during metamorphosis.
“The gene that was disrupted in these mutants was present in all plants, fungi, and animals, including humans, but hadn’t been characterized at all,” Neuman says. She named the gene “Hobbit” due to the small stature of the mutant flies.
She focused her PhD work on figuring out why the mutant animals had such small bodies. Through careful investigation, she identified a likely role for Hobbit in shuttling lipids within cells, a function that was necessary for the release of important growth hormones. Now, after careful analysis of the structure of the Hobbit protein, Neuman and her colleagues have identified a novel superfamily of related proteins and are finding striking links between the proteins’ shape and what they do — including possible links to disease.
A transportation problem
Although lipids play numerous critical roles in cells, moving them around the water-filled spaces is a challenge.
“Lipids are fats, which are hydrophobic, so they don’t play nice with water,” explains Neuman, now a postdoctoral scientist in the Bashirullah Lab. “They need to be kept sequestered from the aqueous environment of the cytoplasm.”
One way to accomplish that is to package up lipids in membrane-bound organelles called vesicles, which can be sent from one place to another within a cell. But that process is slow and fairly resource-intensive — almost like building a new barge each time you want to deliver cargo.
Neuman’s clues pointed to Hobbit functioning as a lipid transfer protein. Like vesicles, lipid transfer proteins move lipids within cells; however, unlike vesicles, lipid transfer proteins accept lipids from one cellular structure and directly hand them off to another, acting more like a cable ferry than a barge.
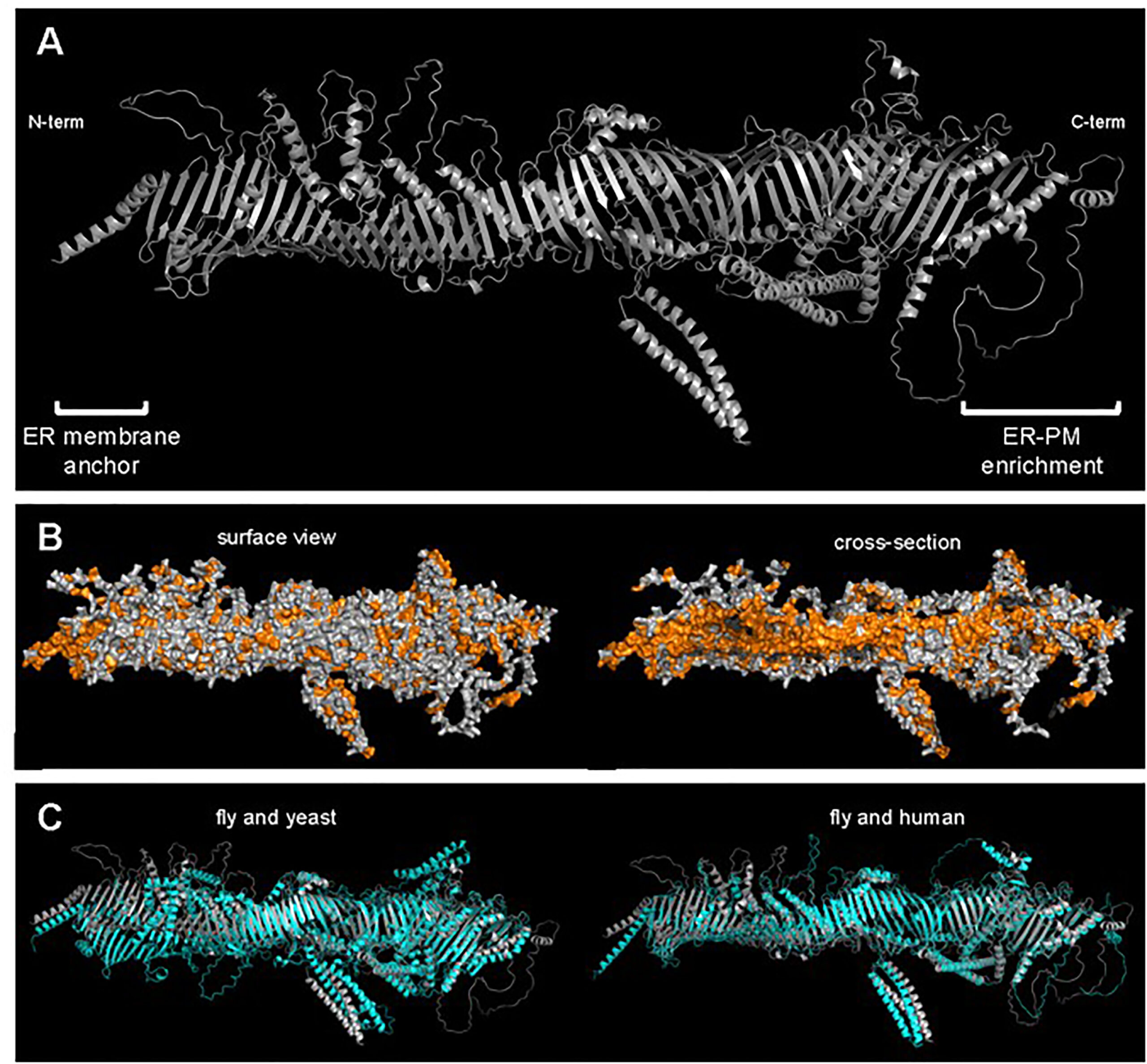
To learn more about how Hobbit transferred lipids, Neuman wanted to know what the protein looked like. Seeing the 3-D structure of a protein is a challenging problem in biology, especially for very large proteins like Hobbit. However, the recent release of an advanced program called AlphaFold provided a new way to solve this problem.
AlphaFold is an algorithm that uses artificial intelligence to predict 3-D protein structures based on their amino acid sequence. In July 2021, AlphaFold’s creators released predicted structures for essentially every protein in the human proteome and the proteomes of several research organisms, including fruit flies. Neuman’s team hurried to examine AlphaFold’s predicted Hobbit structure and were excited at what they saw.
“Basically, the predicted structure of Hobbit looks exactly like what one would expect of a lipid transfer protein,” Neuman says. “The protein literally looks like a tube. If you rotate it so that you’re looking down it, you can see there’s a channel down the very middle. And it just so happens that that channel is lined with hydrophobic amino acids.”
That hydrophobic region keeps the lipids soluble, allowing them to move through the surrounding watery environment.
Neuman, Bashirullah, and collaborator Tim Levine at University College London went looking for other similar proteins in animal genomes and found a total of five different proteins with similar hydrophobic tube shapes. They named the family the bridge-like lipid transfer proteins, or BLTPs, which they described this year in a paper in Trends in Cell Biology.
Building bridges
The elongated channel shape of the BLTPs is very different from other known lipid transfer proteins, most of which look more like a box with a lid, Neuman says, and can hand off a single lipid molecule from one membrane to another. The channel shape, by contrast, is more like a firehose, which might make BLTPs well suited for processes that require moving large amounts of lipids in a short amount of time. In fact, the discovery of this family of proteins provides a possible answer to the long-asked question of how cells rapidly build new membranes.

The researchers still have a lot of questions about which lipids move along the bridge protein and how the flow is regulated. They are particularly intrigued by a modular unit they identified that forms the characteristic hydrophobic groove in all the bridge-like lipid transfer proteins. Each of the five BLTPs has a different number of these repeating beta groove or RBG domains (named in honor of the late Justice Ruth Bader Ginsburg), giving each protein a different tube length.
Each RBG domain also contains a floppier, more variable loop of amino acids, which could be important for regulating protein function, Neuman suggests. For example, if a loop collapsed over the groove, it could act like a physical gate to block or control lipid flow.
The researchers are now planning studies to further explore how the Hobbit protein works and to understand its role during animal development. Mutations in several of the BLTPs are associated with human neurodegenerative and neurodevelopmental disorders, including Alzheimer’s and Parkinson’s diseases, Neuman notes, but it’s still unclear how defects in lipid transfer lead to such diseases.
“The molecular function of Hobbit and the other BLTPs is in transferring lipids between cellular membranes,” she says. “What we don’t yet know, and what we are continuing to study, is how defects in that molecular function lead to human disease.”