
By Eric Hamilton
This article originally appeared on UW–Madison’s news page.
When a patient arrives at a hospital with an infection, her doctor must decide which antibiotic might have the best chance of curing her — no easy feat when disease-causing pathogens are increasingly resistant to multiple antibiotics.
For that reason, hospitals often track the antibiotic resistance profiles of infectious microbes that they isolate from sick patients, which provide information on the most and least effective drugs. But that data is often isolated to a particular hospital and may be difficult to access or hard to interpret, leaving physicians in the dark about which drug is ideal for their patients.
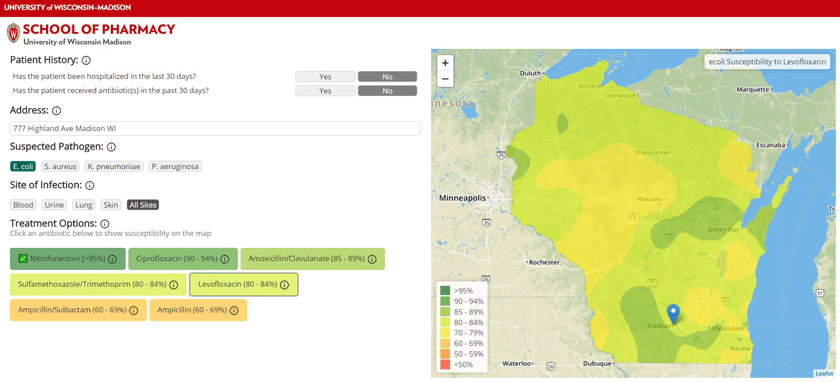
To make this data more accessible, a team of researchers at the University of Wisconsin–Madison School of Pharmacy and the State Cartographer’s Office have developed a prototype system that maps out trends in antibiotic resistance across Wisconsin. They drew inspiration from easy-to-read weather maps and consulted with doctors to develop the tool, which provides guidance at a glance of the likelihood a pathogen will respond to a particular drug.
Having now created a proof-of-concept visualization, the researchers are pursuing finer-grained location data while partnering with hospitals to determine how their new tool, named AMR Tracker, can be incorporated into physician workflows in the future. The researchers published their work online April 1 in the journal Antimicrobial Agents and Chemotherapy.
“With antibiotic resistance, we’ve seen this problem grow very slowly over the past century, and it’s an enormous threat,” says Laurel Legenza (PharmD ’13), an assistant scientist and interim director of global health at the UW–Madison School of Pharmacy, who led the work. “We know there’s an opportunity to improve by selecting an antibiotic that considers the local antibiotic resistance trend. And that’s really the heart of the project, is to provide data in a visualization that would improve its use and application directly to patient care.”
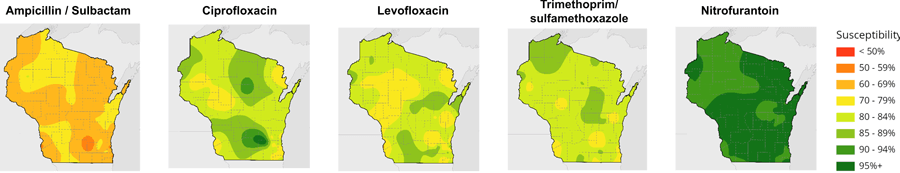
The research team analyzed 202 antibiograms — tables of antibiotic resistance — from Wisconsin hospitals collected between 2009 and 2015, which documented the susceptibility of 200,000 strains of the pathogenic bacterium Escherichia coli to seven classes or combinations of drugs. Plotted on a map of the state, the data created discrete points that then had to be stitched together to produce broad regions of antibiotic susceptibility using cartographic methods contributed by Jim Lacy, Codie See and others at the State Cartographer’s Office.
“It’s incredible what two disciplines can do together that otherwise they wouldn’t do on their own,” Legenza says of the collaboration between pharmacists and cartographers. Associate Professor Susanne Barnett and Associate Professor Warren Rose also guided the project.
The resulting maps reveal significant variation geographically across the state. Resistance to antibiotics tended to be greater in denser, more urban regions of the state as well as in valleys. It’s not clear what underlies these pockets of antibiotic resistance, says Legenza, but her team is investigating factors like population density, antibiotic use patterns, and animal agriculture, which accounts for a sizable share of antibiotic use, to explain the variation. The maps also show that a large proportion of E. coli isolated from patients across much of the state are increasingly resistant to certain antibiotics, like levofloxacin.
In addition to statewide visualizations of susceptibility to a particular drug, AMR Tracker allows users to select a location and see antibiotics ordered by their likelihood of working. The information is color coded: greens for high susceptibility, yellows, oranges and reds for greater resistance.
The researchers have received approval to begin analyzing privacy-protected data from individual patients, which will allow them to pinpoint the location of resistant pathogens more accurately, to individual census tracts. They are also partnering with the Marshfield Clinic Health System to collect genomic data from antibiotic-resistant microbes to improve the maps.
Incorporating a tool like AMR Tracker into the health care system may be years away. Next steps include validating the geographic smoothing of the data, including additional pathogens and working with hospital systems and electronic medical records companies to integrate AMR Tracker into patient visits.
“The more seamless we can make this, the greater value it will offer to providers and their patients,” says Legenza.
Learn how the AMR Tracker got its start.