
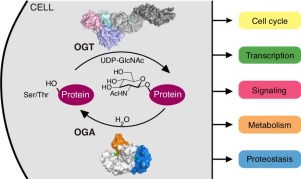
The first and most essential steps for decoding the complex roles of O-GlcNAcylation in cells is gaining a detailed understanding of the enzymes that regulate this modification. OGT and OGA are a single pair of human enzymes that catalyze the reversible O-GlcNAcylation of more than 4,000 proteins. Interestingly, no apparent recognition motif has been identified in O-GlcNAc modification sites, making it unclear how OGT and OGA systematically discriminate their substrates. Further, it is known that both OGT and OGA associate with many non-substrate proteins in cells, though their mechanisms of interaction are poorly understood. Structural Biology has been the key to unlocking much of what we currently know about the OGT and OGA catalytic mechanisms and substrate recognition (see Selected Publication #1 below). In a major advance for the field, we recently solved the crystal structure of a human OGA construct, representing the first model of a eukaryotic glycoside hydrolase in the GH84 family (see Selected Publication #2 below). We further determined a series of structures of OGA in complex with a small molecule inhibitor, as well as various O-GlcNAcylated substrates (see Selected Publication #3 below). These studies have both confirmed the previously proposed hydrolytic mechanism of OGA and elucidated novel modes of substrate binding. Work in this area continues to push forward our understanding of the OGT and OGA structures and the unique features that allow them to coordinate and communicate across the cell. We expect these efforts to lay the foundation for understanding O-GlcNAc biology and development of OGT and OGA as new drug targets.
Selected Publications
- Joiner CM, Li H, Jiang J*, Walker S*. Structural characterization of the O-GlcNAc cycling enzymes: insights into substrate recognition and catalytic mechanisms. Current Opinion in Structural Biology (2019) 56, 97-106.
- Li B, Li H, Lu L, Jiang J*. Structures of human O-GlcNAcase and its complexes reveal a new substrate recognition mode. Nature Structural & Molecular Biology (2017), 24, 362-369. [News and Views, Nat Struct Mol Biol. 2017, 24, 433.]

- Li B#, Li H#, Hu CW, Jiang J*. Structural insights into the substrate binding adaptability and specificity of human O-GlcNAcase. Nature Communications (2017) 8:666. doi: 10.1038/s41467-017-00865-1. (#equal contribution)
