Skip to main content
- Hong ZZ, Lowe J, Jiang J*. Dissecting the mechanisms underlying the substrate recognition and functional regulation of O-GlcNAc cycling enzymes. ACS Chemical Biology. 2025, 20, 2534-2546.
- Wang A#, Young M#, Jiang J*. The Glyco-Switch of life: O-GlcNAcylation in cell fate decision. Glycobiology. 2025, 35, cwaf061. (#equal contribution)
- Sun F, Wang K, Dong X, Secaira-Morocho H, Hui A, Cai C, Sze J, Low B, Udgata S, Pasch C, Huan T, Deming D, Zhu Q, Jiang J, Fu T. The microbial bile acid metabolite 3-oxo-LCA inhibits colorectal cancer progression. Cancer Research. 2025, 85(24), 4937-4957.
- Dong X, Sun F, Secaira-Morocho H, Hui A, Wang K, Cai C, Udgata S, Low B, Wei S, Chen X, Qi M, Pasch CA, Xu W, Jiang J, Zhu Q, Huan T, Deming DA, Fu T. The dichotomous roles of microbial-modified bile acids 7-oxo-DCA and isoDCA in intestinal tumorigenesis. Proceedings of the National Academy of Sciences of the United States of America. 2024, 121(47): e2317596121.
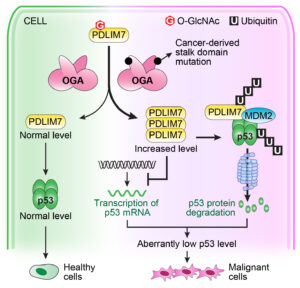
- Hu CW, Wang A, Fan D, Worth M, Chen Z, Huang J, Xie J, Macdonald J, Li L, Jiang J*. OGA mutant aberrantly hydrolyzes O-GlcNAc modification from PDLIM7 to modulate p53 and cytoskeleton in promoting cancer cell malignancy. Proceedings of the National Academy of Sciences of the United States of America. 2024, 121(24): e2320867121. Previous preprint of the manuscript: https://doi.org/10.21203/rs.3.rs-2709128/v1.
- Hu CW, Wang K, Jiang J*. The non-catalytic domains of O-GlcNAc cycling enzymes present new opportunities for function-specific control. Current Opinion in Chemical Biology. 2024, 81: 102476.
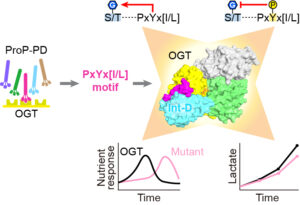
- Blankenship CM#, Xie J#, Benz C, Wang A, Ivarsson Y, Jiang J*. Motif-dependent binding on the intervening domain regulates O-GlcNAc transferase. Nature Chemical Biology. 2023, 19(11): 1423-1431. Previous preprint of the manuscript: https://doi.org/10.21203/rs.3.rs-2531412/v1. (#equal contribution)
- Hu CW, Xie J, Jiang J*. The emerging roles of protein interactions with O-GlcNAc cycling enzymes in cancer. Cancers. 2022, 14(20), 5135. https://doi.org/10.3390/cancers14205135.
- Jiang J*, Williams SJ*. Editorial overview: Protein-carbohydrate complexes and glycosylation: A new age of discovery in the glycosciences. Current Opinion in Structural Biology. 2021 Jun;68:iii-v. doi: 10.1016/j.sbi.2021.03.002.
- Kositzke A#, Fan D#, Wang A, Li H, Worth M, Jiang J*. Elucidating the protein substrate recognition of O-GlcNAc transferase (OGT) toward O-GlcNAcase (OGA) using a GlcNAc electrophilic probe. International Journal of Biological Macromolecules. 2020 Dec 18;169:51-59. doi: 10.1016/j.ijbiomac.2020.12.078. Online ahead of print. (#equal contribution)

- Blankenship C#, Xie J#, Jiang J*. Nucleocytoplasmic protein glycosylation. Comprehensive Glycoscience, 2nd edition. Available online 30 November 2020. (#equal contribution)
- Estevez A#, Zhu D#, Blankenship C, Jiang J*. Molecular interrogation to crack the case of O-GlcNAc. Chemistry–A European Journal. (2020) 26, 12086-12100. (#equal contribution)

- Worth M#, Hu CW#, Li H, Fan D, Estevez A, Zhu D, Wang A, Jiang J*. Targeted covalent inhibition of O-GlcNAc transferase in cells. Chemical Communications (2019) 55, 13291-4. (#equal contribution)

- Joiner CM, Li H, Jiang J*, Walker S*. Structural characterization of the O-GlcNAc cycling enzymes: insights into substrate recognition and catalytic mechanisms. Current Opinion in Structural Biology (2019) 56, 97-106.
- Hu CW, Worth M, Li H, Jiang J*. Chemical and biochemical strategies to explore the substrate recognition of O-GlcNAc-cycling enzymes. Chembiochem (2019) 20, 312-8.

- Hu CW#, Worth M#, Fan D#, Li B#, Li H#, Lu L, Zhong X, Lin Z, Wei L, Ge Y, Li L, Jiang J*. Electrophilic probes for deciphering substrate recognition by O-GlcNAc transferase. Nature Chemical Biology (2017) 13, 1267-73. (#equal contribution)

- Liu F, Ma F, Wang Y, Hao L, Zeng H, Jia C, Wang Y, Liu P, Ong IM, Li B, Chen G, Jiang J, Gong S, Li L, Xu W. PKM2 methylation by CARM1 activates aerobic glycolysis to promote tumorigenesis. Nature Cell Biology (2017) 19, 1358-70.
- Li B#, Li H#, Hu CW, Jiang J*. Structural insights into the substrate binding adaptability and specificity of human O-GlcNAcase. Nature Communications (2017) 8:666. doi: 10.1038/s41467-017-00865-1. (#equal contribution)

- Li B, Li H, Lu L, Jiang J*. Structures of human O-GlcNAcase and its complexes reveal a new substrate recognition mode. Nature Structural & Molecular Biology (2017), 24, 362-369. [News and Views, Nat Struct Mol Biol. 2017, 24, 433.]

- Worth M#, Li H#, Jiang J*. Deciphering the functions of protein O-GlcNAcylation with chemistry. ACS Chemical Biology (2017), 12, 326-335. (#equal contribution)

- Lu L, Fan D, Hu CW, Worth M, Ma ZX, Jiang J*. Distributive O-GlcNAcylation on the highly repetitive C-terminal domain of RNA polymerase II. Biochemistry (2016), 55, 1149-1158.

- Ortiz-Meoz RF, Jiang J, Lazarus MB, Orman M, Janetzko J, Fan C, Duveau DY, Tan ZW, Thomas CJ, Walker S. A small molecule that inhibits OGT activity in cells. ACS Chemical Biology (2015), 10, 1392-1397.
- Lazarus MB*, Jiang J*, Kapuria V, Bhuiyan T, Janetzko J, Zandberg WF, Vocadlo DJ, Herr W, Walker S. HCF-1 is cleaved in the active site of O-GlcNAc transferase. (*equal contribution) Science (2013), 342, 1235-1239.
- Lazarus, MB*; Jiang, J*; Gloster, TM; Zandberg, WF; Vocadlo, DJ; Walker, S. Structural snapshots of the reaction coordinate for O-GlcNAc transferase. (*equal contribution) Nature Chemical Biology (2012), 8, 966-968.
- Jiang, J*; Lazarus, MB*; Pasquina, L; Sliz, P; Walker, S. A neutral diphosphate mimic crosslinks the active site of human O-GlcNAc transferase. (*equal contribution) Nature Chemical Biology (2012), 8, 72–77.
- Lazarus, MB; Nam, Y; Jiang, J; Sliz, P; Walker, S. Structure of human O-GlcNAc transferase and its complex with a peptide substrate. Nature (2011), 469, 564-567.
- Jiang, J; Tetzlaff, CN; Takamatsu, S; Iwatsuki, M; Komatsu, M; Ikeda, H; Cane, DE. Genome mining in Streptomyces avermitilis. A biochemical Baeyer-Villiger reaction and discovery of a new branch of the pentalenolactone family tree. Biochemistry (2009), 48, 6431-6440.
- Giglio, S; Jiang, J; Saint, CP; Cane, DE; Monis, PT. Isolation and characterization of the gene associated with geosmin production in cyanobacteria. Environmental Science & Technology (2008), 42, 8027-8032.
- Jiang, J; Cane, DE. Geosmin biosynthesis. Mechanism of the fragmentation-rearrangement in the conversion of germacradienol to geosmin. Journal of the American Chemical Society (2008), 130, 428-429.
- Nawrath, T; Dickschat, JS; Muller, R; Jiang, J; Cane, DE; Schulz, S. Identification of (8S,9S,10S)-8,10-dimethyl-1-octalin, a key intermediate in the biosynthesis of geosmin in bacteria. Journal of the American Chemical Society (2008), 130, 430-431.
- Vedula, SL; Jiang, J; Zakharian, T; Cane, DE; Christianson, DW. Structural and mechanistic analysis of trichodiene synthase using site-directed mutagenesis: probing the catalytic function of tyrosine-295 and the asparagine-225/serine-229/glutamate-233-Mg2+B motif. Archives of Biochemistry and Biophysics (2008), 469, 184-194.
- Jiang, J; He, X; Cane, DE. Biosynthesis of the earthy odorant geosmin by a bifunctional Streptomyces coelicolor enzyme. Nature Chemical Biology (2007), 3, 711-715.
- Jiang, J; He, X; Cane, DE. Geosmin biosynthesis. Streptomyces coelicolor germacradienol/germacrene D synthase converts farnesyl diphosphate to geosmin. Journal of the American Chemical Society (2006), 128, 8128-8129.
- Zhou, C; Jiang, J; Zhou, Y; Xie, Z; Miao, Q; Wang, Z. Chemoselective carbonyl benzylation mediated by Zn/CdCl2/InCl3 in tap water. Letters in Organic Chemistry (2005), 2, 61-64.
- Zha, Z; Qiao, S; Jiang, J; Wang, Y; Miao, Q; Wang, Z. Barbier-type reaction mediated with tin nano-particles in water. Tetrahedron (2005), 61, 2521-2527.
- Zhou, C; Zhou, Y; Jiang, J; Xie, Z; Wang, Z; Zhang, J; Wu, J; Yin, H. Organometallic reactions in aqueous media: the allylations of carbonyl compounds mediated in Zn/CdSO4 and Zn/SnCl2 bimetal systems. Tetrahedron Letters (2004), 45, 5537-5540.













